-
background
The knowledge about the ion channels and transporters on the cell membrane are rapidly accumulating, and now, we can construct various quantitative cell models based on these findings. In our laboratory, we are trying to construct quantitative models of cells and orgrans, especially about heart.
These models are basically formalized as mathematical equations, and many cellular or subcellular models are now being developed. However, the model research concerning organ level systems are still limited, for example heart and other organ models developed in Auckland University group in New Zealand. Such research area is now called Physiome area, which is one of the post genomic projects.
These organ level quantitative models are useful to understand complex phenomena in whole body. And also, these models may become powerful tools in the clinical applications, since they can be a testbed for certain treatments, and also a testbed for drug discovery.
-
heart simulation
The main research topic in our laboratory is construction of heart model which can reproduce physiological hemodynamics based on the detailed cell model which has almost all known ion channels, transporters, contraction proteins and organelles.
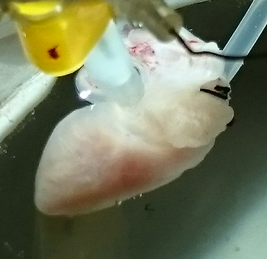
Cells including ventricular myocytes, have special ion transportation system such as ion channels and ion transporters on their membranes and ions move according to the intracellular and extracellular ion concentrantions or membrane potentials or other factors. For the ventricular myocytes, there are special calcium buffering system called sarcoplasmic reticulum which stores and releases calcium and actin and myosin filaments which develop contraction force. Functions of almost all of these intracellular proteins are now described as a set of ordinary differential equations (ODEs). For example, recent cardiac cell model Kyoto Model has about 50 ODEs.

By connecting 100 millions of cardiac cell models, we can construct a heart model. However, there is still many unknown mechanism of Heart, thus some of the physiological properties cannot be reproduced by such simple model. For example, although a ventricular myocyte shortens for about 15% of its length, heart pumps out 60 – 70 % of its blood in a heart beat. In real heart, the tissue has sheet structure and they slip each other which enables large deformation with small length change. Also the fiber orientation of heart wall is known to have variation from epicardium to endocardium and this makes rotation at heart beat. However, the detailed mechanism of such deformation is still unknown thus our goal is to make hypothesis of how the heart is constructed to achieve high pump performance.
-
retinal cell model
Visual system is one of the main sensors for human to understand environments. Thus the damage in vision dramatically lowers QOL. In our laboratory, we also are trying to develop quantitative cellular models in retina. Since many of the retinal diseases are related with the imbalance of energy production and demand, our models are aimed to reproduce the energy demand in these cells.
The primary model that we focused on is the photoelectric transducer system in photoreceptor cells. Detailed experimental data are measured by Dr. Kawamura, former professor of Osaka University, and we are collaborating to construct accurate phototransduction system. By incorporating this system, we are also developing accurate electrophysiological model of photoreceptor cells which include known ion channels and transporters in photoreceptor cells.
Finally, we are planning to make a retinal model which can reproduce ERG (Electro Retinogram) which is widely used in the diagnose in retinal diseases.
-
electronic text book e-Heart project
In the project conducted by the Biosimulation Research Center in Ritsumeikan University, electronic textbook called e-Heart is developed. The project is conducted by Prof. Akinori Noma, and the contents of the textbook is based on the simulation software for various cell types, phenomena and orgrans. Current version contains following topics.
- human ventricular myocyte model
- SA node pacemaker cell model
- atrial myocyte model
- electrical circuit model of cell membrane
- simplified action potential model
- single K channel model
- sing Ca channel model
- NaK pump model
- NaCa exchanger model
- excitation propagation model
- excitation propagation with gapjunction model
- excitation propagation model in 2D sheet
- abnormal excitation propagation
- simplified circulation model
- capillary model
- mitochondria model
- enzyme reaction
- competitive and non-competitive inhibition in enzyme reaction
- muscle contraction element model
- ion concentration homeostasis model